Using the E.coli JM109 genome as a template, using pQE-nlpE-F and pQE-nlpE-R, pQE-spy-F and pQE-spy-R as primers respectively, 2 genes were amplified through high-fidelity PCR Fragments, sizes were 711 bp and 486 bp respectively. PCR program: 95°C for 5 min; 98°C for 10 s, 55°C for 15 s, 72°C for 1 min, 30 cycles; 72°C for 10 min, 16°C for 30 s.
The DNA polymerase SYBR® Premix Ex Taq Ⅱ (Tli RNaseH Plus) (2×Conc.), high-fidelity DNA polymerase PrimerSTAR® and restriction endonucleases used in the real-time fluorescence quantitative PCR process were all purchased from Dalian Bao Biotechnology Co., Ltd., one-step cloning ligase Exnase Ⅱ was purchased from Nanjing Novezan Biotechnology Co., Ltd., agarose gel DNA recovery kit, plasmid miniprep kit and PCR product purification kit were purchased from Shanghai Jierui Bioengineering Co., Ltd., other materials and reagents used are of domestic analytical grade. Primers were synthesized by Suzhou Hongxun Biotechnology Co., Ltd., and gene sequencing services were provided by Saiyin Biotechnology (Shanghai) Co., Ltd. LB medium: 0.5% yeast extract, 1.0% tryptone, 1.0% NaCl, sterilized at 1×105 Pa for 20 min.
DNA preparation: Transfer the overnight activated E. coli JM109 to 30 mL of fresh LB liquid medium at an inoculation volume of 1% (V/V), and culture at 37°C and 120 r/min until the OD600 is approximately When the value is 0.8, add different concentrations of butanol (0.2%–0.8%, V/V) to the shake flask. After continuing to incubate for 90 minutes, absorb 2 mL of bacterial liquid, centrifuge at 5510 × g for 2 minutes at room temperature, and pour out the supernatant. The bacterial cells were collected from the culture medium and RNA was extracted. The corresponding cDNA was synthesized using a one-step cDNA synthesis (reverse transcription) kit (Shanghai Jierui Bioengineering Co., Ltd.).
Fluorescence quantitative PCR: Use the above cDNA as a template, qPCR-rcsB-F/R, qPCR-cpxR-F/R, qPCR-baeR-F/R, qPCR-pspA-F/R, qPCR-rpoE- F/R is the primer (Table 1), using the SYBR® Green Ⅰ chimeric fluorescence method (Dalian Bao Bioengineering Co., Ltd.). SYBR® Green Ⅰ emits fluorescence after combining with double-stranded DNA, and the double-stranded DNA is generated by detecting the PCR reaction. SYBR®Green Ⅰ combines the fluorescence intensity emitted to achieve the purpose of accurate quantification of the target gene. Please refer to the kit instructions for the reaction system.

Use two restriction endonucleases, BamH Ⅰ and Hind Ⅲ, to linearize the pQE80L vector by double digestion. The digestion conditions are 37°C for 6–8 h.
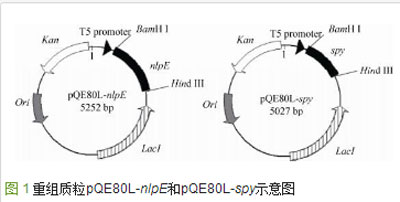
The nlpE and spy gene fragments amplified by PCR with homology arms were ligated with the pQE80L vector double-digested by BamH Ⅰ and Hind Ⅲ restriction enzymes through a one-step cloning enzyme method (Nanjing Novell Zan Biotechnology Co., Ltd.) were connected to construct plasmids pQE80L-nlpE and pQE80L-spy (Figure 1). The one-step cloning and ligation system is: 50-200 ng linearized pQE80L, 20-200 ng insert fragment, 2 μL 5×CE Ⅱ Buffer, 1 μL Exnase Ⅱ. The ligation system was placed at 37°C for 30 min to react. After the reaction was completed, it was cooled on ice for 5 min. The ligation product was transferred into E. coli JM109 competent cells through chemical transformation and spread on a solid containing kanamycin. flat. The next day, the positive single colony on the plate was preliminarily verified by colony PCR, and the verified positive transformants were sequenced and verified.
Use spy-K-F/R and nlpE-K-F/R as primers respectively, and use plasmid pKD13 as template to amplify the FRT-Kan-FRT fragment (about 1450 bp) with homology arms. The source arm FRT-Kan-FRT fragment was transferred into the competent E. coli JM109 containing pKD46 and integrated into the E. coli JM109 genome. The successfully verified positive recombinant was prepared into competent cells according to the preparation method of chemically transformed competent cells, and the plasmid pCP20 was electrotransduced into the competent cells, cultured at 30°C overnight, and then cultured at 42°C for 10 h. Eliminate plasmid pCP20.
Transfer the overnight-activated recombinant bacteria into 30 mL of fresh LB liquid medium at an inoculum volume of 1% (V/V), and culture the cells at 37°C and 120 r/min until the OD600 reaches 0.6, add IPTG with a final concentration of 0.2 mmol/L was induced and cultured at 30°C for 6 h, and the cells were collected by centrifugation at 8801×g for 10 min at 4°C. Ultrasonic disruption was carried out on an ultrasonic disruptor for 15 min (ultrasonic 1 s, 3 s interval, 500 W), and the disrupted liquid was subjected to SDS-PAGE to analyze protein expression.
Culture the recombinant bacteria and E. coli JM109 (control bacteria) with pQE80L through transfer induction (0.2 mmol/L IPTG) until the OD600 reaches 0.8, add 0.8% (V/V) n-butanol into the cell culture medium and cultured at 30°C and 120 r/min for 10 h. During this period, samples were taken every 2 h and the growth status of the bacteria was monitored through the OD600 reading of the spectrophotometer.
The microbial adhesion to hydrocarbons (MATH) method was used to measure changes in cell surface hydrophobicity. According to the method of measuring the tolerance of recombinant bacteria, the control strain E. coli JM109/pQE80L and the recombinant strains E. coli JM109/pQE80L-nlpE and E. coli JM109/pQE80L-spy after overnight activation were induced and cultured by transfer (0.2 mmol/L IPTG) for 6 h, and centrifuge at 4°C and 8801×g for 10 min to collect the cells. Aspirate the culture medium thoroughly, resuspend the cells in 0.1 mol/L potassium phosphate buffer (pH 6.0), and control the initial OD400 value to 0.8–0.9 (A0). Take 4.8 mL of bacterial liquid and 0.8 mL of solvent, mix thoroughly, let stand at room temperature for 15 minutes, take a water phase sample after layering and measure its OD400 value (A1). The adhesion rate is calculated using formula (1).
Microbial culture media products for molecular biology and protein expression:
▼▲
English name
Chinese name
CAS code number
ONPG
2-nitrobenzeneβ-D-Galactopyranoside [Medium for β-D-galactose]
369-07-3
GPNPG
4-Nitrophenyl-β-D-galactopyranoside [Medium for β-galactose]
3150-24-1
D-(-)-Luciferin [Chemiluminescence Reagent]
D-(-)-fluorescein [chemiluminescence reagent]
2591-17-5
X-Gal
5-Bromo-4-chloro-3-indolyl β-D-galactanthracene [for biochemical research]
7240-90-6
Magenta-Gal (contains ca. 10% Ethyl Acetate)
5-Bromo-6-chloro-3-indolyl-β-D-galactanthracene (containing approximately 10% ethyl acetate) [for biochemical research]
93863-88-8
G Bluo-Gal
5-Bromo-3-indolyl-β-D-galactanthracene [for biochemical research]
97753-82-7
G Rose-Gal
6-Chloro-3-indole-β-D-galactanthracene [for biochemical research]
138182-21-5



 微信扫一扫打赏
微信扫一扫打赏
